This article has been reviewed according to Science X'seditorial processandpolicies.Editors强调了以下属性而在suring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Optimizing sepsis treatment timing with a machine learning model
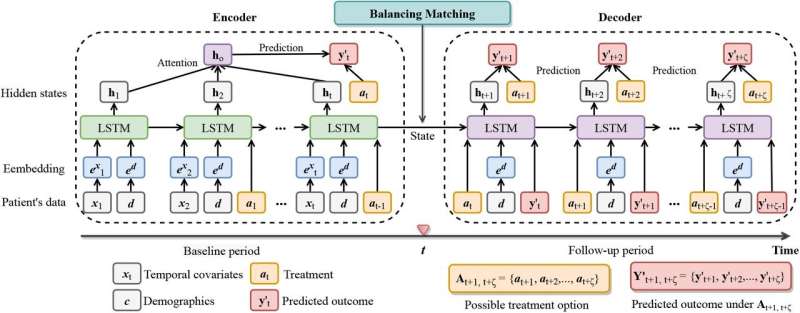
A new machine learning model that estimates optimal treatment timing for sepsis could pave the way for support tools that help physicians personalize treatment decisions at the patient bedside, researchers say.
In a paper published inNature Machine Intelligence, scientists from The Ohio State University describe the new model, which uses artificial intelligence to take on the complex question of when to administer antibiotics to patients with a suspected case of sepsis.
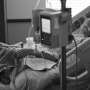
Time is of the essence because sepsis, the body's overwhelming response to an infection, can rapidly lead to organ failure. And yet, its symptoms—fever,low blood pressure, increasingheart rateand breathing problems—can look like a lot of other conditions. Federal guidelines call for quick treatment withbroad-spectrum antibioticsas the first line of defense—a strategy that typically requires action before cultures confirming a bacterial infection can be obtained from a lab.
The model was designed to take into account these uncertainties and time pressures.
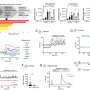
Researchers tested the model's performance using critical-carepatient informationfrom a U.S. database and a European database, comparing outcomes in patients whose actual treatment matched the model's recommended treatment timeline to outcomes for patients whose actual treatment had differed from what the model would have recommended based on their vital signs, lab results and risk-related demographic data. The measure representing the outcome was patient survival 30 and 60 days after sepsis treatment.
"We showed that when the actual treatment andartificial intelligenceagree, we have a lower mortality rate. If they don't agree, the mortality rate can be as high as 25%," said senior author Ping Zhang, Ph.D., assistant professor of computer science and engineering and biomedical informatics at Ohio State.
The model was trained and validated on a dataset obtained from a publicly available database, calledMIMIC-III. The model was tested on different portions of MIMIC-III and a new external dataset fromAmsterdamUMCdb.
Key measures from almost 14,000 individuals with sepsis included changes to patientvital signsand lab test results as time passed—serving as indicators of illness severity and type of infection—and an innovative method devised to compare outcomes for patients who did and did not receive antibiotics at a specific time.
"We want the modeling to predict whether it's beneficial to use antibiotics at a given time—yes or no. But we'll never know what happens if we don't give the antibiotic. So we applied a clinical trial concept to this model: For every patient who had taken the drug, we included a matched, clinically similar patient who didn't take antibiotics at that time," said Zhang, who leads the Artificial Intelligence in Medicine Lab and is also a core faculty member in Ohio State's Translational Data Analytics Institute.
"That way, we can predict the counterfactual outcome, and train the counterfactual treatment model to find whether treatment for sepsis works or not."
脓毒症导致的三分之一以上spital deaths, and is seen most often in intensive care units and emergency departments, "where we're often making decisions without the gold standard—results from a culture," said study co-author Katherine Buck, MD, assistant professor of emergency medicine in the College of Medicine and director of the Geriatric Emergency Department at Ohio State Wexner Medical Center. "Not every patient that meets sepsis criteria goes on to have proof of a bacterial infection."
Antibiotics don't come without risks—they can be toxic to kidneys, prompt an allergic reaction or lead to C. difficile, an infection that causes severe diarrhea and inflammation of the colon.
"What this paper starts to get at is, can we use information available to the clinicians, sometimes at the forefront and sometimes not, to say, Things are changing in a way that suggests the patient will benefit from antibiotics," Buck said. "A decision-support tool could tell clinicians if it matches what we're already thinking or prompt us to ask ourselves what we're missing. Hopefully, with time, all the electronic health record data we have will reveal signals—and from there it's a matter of figuring out how to use them and how to get that to clinicians."
Those insights—and availability of electronic health record data—were important to feeding the model with the right kind of data and designing it to take into account multiple considerations that come with changing medical circumstances, Zhang said.
"We modeled the patient record like it's language," he said. "And formachine learning, we always train the model batch by batch—you need the model to analyze the pattern of data, set parameters, and based on these parameters, add another training dataset to make improvements. And then the machine always finds better parameters to fit the model."
A key measure used to guide how the model arrives at a recommendation is the Sequential Organ Failure Assessment (SOFA) score, which is used to regularly assess how an ICU patient's organ systems are performing based on results from six lab tests. The researchers ran example case studies to demonstrate what an interface developed for theclinical settingmight look like, showing how SOFA scores change when the model adjusts the recommended treatment timeline based on changes to personalized patient data.
"Our paper is the first to use AI to pursue an antibiotic recommendation for sepsis, using real-world data to help clinical decision making," Zhang said. "Any research like this needs clinical validation—this is phase one for retrospective data analysis, and phase two will involve human-AI collaboration for better patient care."
More information:Ping Zhang, Estimating treatment effects for time-to-treatment antibiotic stewardship in sepsis,Nature Machine Intelligence(2023).DOI: 10.1038/s42256-023-00638-0.www.nature.com/articles/s42256-023-00638-0